WGS
Human
Introduction
Offers genome wide detection of DNA variants including SNP, CNV, Indel and SV
Ability to detect rare and novel (de novo) genetic variants
Applicable to patients whose WES has failed to detect pathogenic variants
Sample type: Genomic DNA
Technology
Wet lab
NovaSeq 6000
Average sequencing depth ≥30X
Dry lab (Bioinformatics)
Case sharing - gastric cancer-related genetic variation information
Gastric cancer is a heterogeneous disease with different molecular etiologies. We performed WGS of 100 pairs of tumor-normal samples at an average effective sequencing depth of 84X.
Bioinformatics analysis identified tumour-specific somatic mutations in known genes such as TP53, ARID1A, CDH1 and in a newly discovered gene MUC6. Mutations in driver genes were also found including CTNNA2, GLI3, and RNF43. The study also revealed that that 4.3% of RHOA mutations in diffuse tumors were closely related to the most common interference pathways (adhesion links and focal adhesions) in gastric cancer.
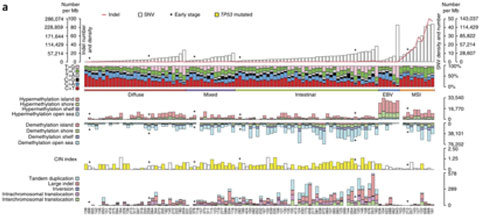
Kai Wang, Siu Tsan Yuen, et al., Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nature Genetics. (2014),doi:10.1038/ng.2983.
Why choose Berry Genomics for WGS
-
Output as high as 200,000 Gb raw data per week
-
Integrated professional teams for sequencing, data analysis and reporting
-
Fast TAT to report of 25 days
-
Comprehensive disease report for clinical geneticist, including supplementary file listing all potentially pathogenic mutations
-
Option of uploading raw data by Cloud to local hospitals for storage and reanalysis
Animal or Plant
Introduction
The genome-wide sequencing of animals and plants genomes is important to understand differences in species and varieties. The information can be used for population genetic polymorphism analysis, evolutionary analysis, identification of new functional genes, breeding guidance and genetic modifications.
Technology
NovaSeq 6000
Obtain information on CDS, Introns, intergenic regions
SNP, CNV, InDel, SV and other variation information
Structural variations and genomic CNVs in large fragments
Case sharing – Genome-wide resequencing for Soybean
Identification of domesticated and improved genes in the soybean genome is important for guiding soybean breeding. Three types of germplasm lines were selected in this study. A total of 302 representative soybean germplasm resources were sequenced at a depth was greater than 11X. The results showed that the genetic polymorphism of soybean was significantly reduced during acclimation and improvement. Genome-wide association analysis on seed size, seed coat color, growth habit, oil content and other traits identified a series of significant associated genetic sites.
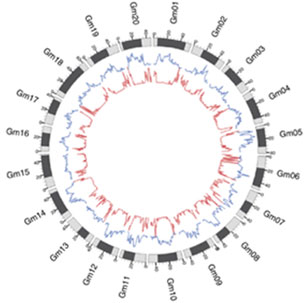
The decline of linkage imbalance (LD) in GWAS analysis of soybean genomic population
Zhou Z, Jiang Y, et al., Resequencing 302 wild and cultivated accessions genes related to domestication and improvement in soybean. Nat Biotechnol.(2015),doi:10.1038/nbt.3096.

